Hello! My name is Nicholas Ho and I am a third year
Computational Biology PhD student in Carnegie Mellon University School of Computer Science.
I am incredibly grateful to be advised by
Professor Jian Ma and
Professor Eric Xing.
I care a lot about building models that can both accurately capture biology and uncover new insights, while genuinely scaling with data and model size.
In other words, I aim to develop truly useful biological foundation models.
In the past, I led the pretraining and evaluation for
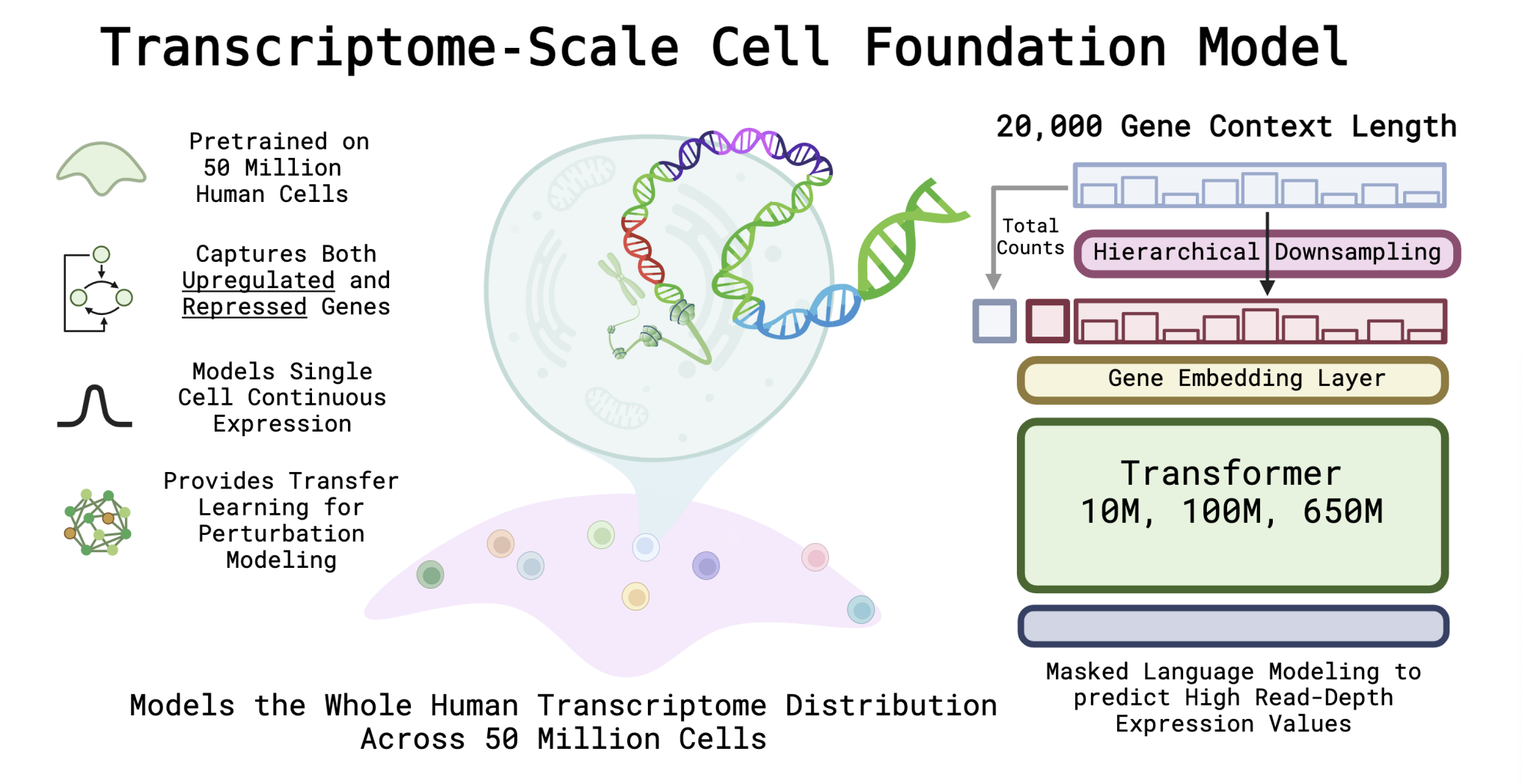
AIDO.Cell (600M Parameter whole transcriptome model for cell) and
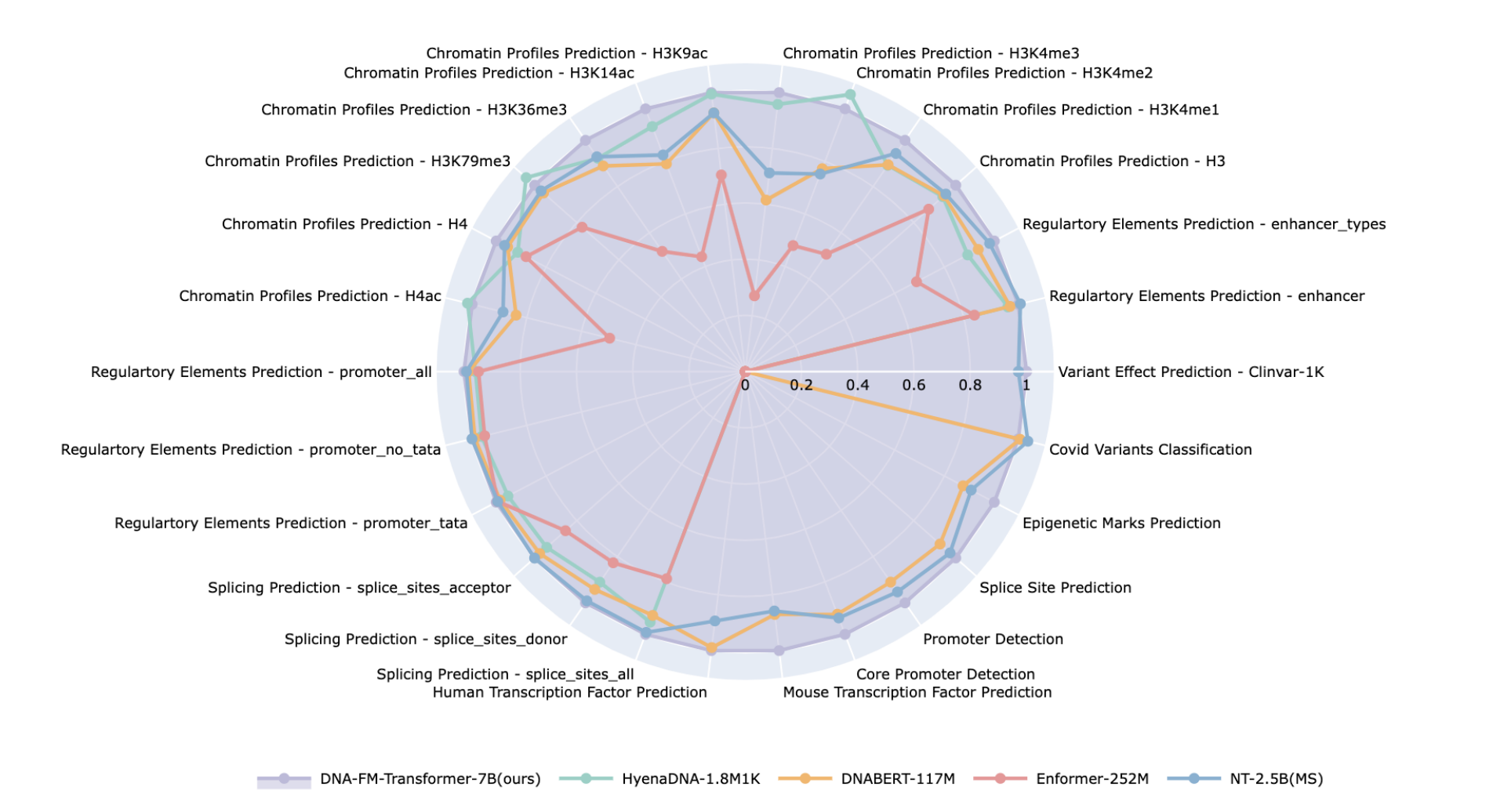
AIDO.DNA (7 Billion Parameter model for DNA) at
GenBio.AI.
While I think there is still a very, very long way to go, I am optimistic about the future for better models in biology.
- (1) 🔬 Single-Cell perturbation modeling, specifically for unseen chemical and genetic perturbations.
- (2) 🧪 Cis-Regulatory sequence to function modeling, more recently, I've pretrained DNA language models to see how evolution can be useful for this.
🤖 As a machine learning scientist, I work on:
- (1) 💻 Training large-scale models, at least Billions of parameters requiring 3D parallelism. I have a great love-hate relationship with NVIDIA's Megatron-LM.
- (2) 🌊 Diffusion & Flow matching models for generative modeling. So far I've used these tools most for single cells. Coming from a biophysics background, this stuff is so cool.
- (3) 📚 Representation learning, either through self-supervision or multi-task learning. In the past, I worked on it for modeling drug representations. Also I am a Big JEPA fan.
Quick Note on Foundation Models
Although I would say I work on "foundation models" (which I think is an immensely overloaded term), I believe that the task always informs the representation. Although this sounds like it may contradict the point of foundation models (where a model may be useful for any task), in biology - because the data is limited, it is important to design the right pretraining objective that is best aligned with the important downstream tasks. By working on the right problems with the right inductive biases, then I believe we can achieve generalizable & scalable models.
Feel free to reach out to me! I'm always looking for interesting problems to talk about!
Publications
Most recent publications on Google Scholar.
‡ indicates equal contribution.
Scaling Dense Representations for Single Cell with Transcriptome-Scale Context
Nicholas Ho, Caleb N. Ellington, Jinyu Hou, Sohan Addagudi, Shentong Mo, Tianhua Tao, Dian Li, Yonghao Zhuang, Hongyi Wang, Xingyi Cheng, Le Song, Eric P. Xing.
In NeurIPS Workshop on AI for New Drug Modalities, 2024.
Accurate and General DNA Representations Emerge from Genome Foundation Models at Scale
Caleb N. Ellington, Ning Sun, Nicholas Ho, Tianhua Tao, Sazan Mahbub, Dian Li, Yonghao Zhuang, Hongyi Wang, Le Song, Eric P. Xing.
In NeurIPS Workshop on AI for New Drug Modalities, 2024.
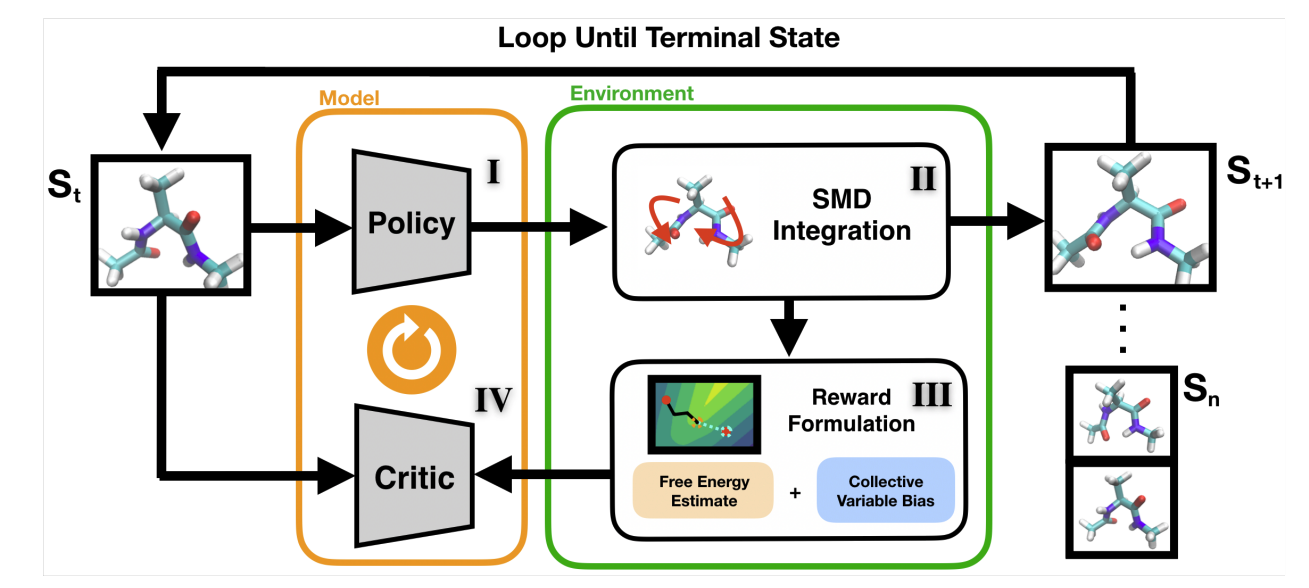
Learning Free Energy Pathways through Reinforcement Learning of Adaptive Steered Molecular Dynamics
Nicholas Ho, John Kevin Cava, John Vant, Ankita Shukla, Jacob Miratsky, Pavan Turaga, Ross Maciejewski, Abhishek Singharoy.
Machine Learning In Structural Biology (MLSB) Workshop at the 36th Conference on Neural Information Processing Systems
Scaling Dense Representations for Single Cell with Transcriptome-Scale Context
Nicholas Ho, Caleb N. Ellington, Jinyu Hou, Sohan Addagudi, Shentong Mo, Tianhua Tao, Dian Li, Yonghao Zhuang, Hongyi Wang, Xingyi Cheng, Le Song, Eric P. Xing.
In NeurIPS Workshop on AI for New Drug Modalities, 2024.
Accurate and General DNA Representations Emerge from Genome Foundation Models at Scale
Caleb N. Ellington, Ning Sun, Nicholas Ho, Tianhua Tao, Sazan Mahbub, Dian Li, Yonghao Zhuang, Hongyi Wang, Le Song, Eric P. Xing.
In NeurIPS Workshop on AI for New Drug Modalities, 2024.
Learning Free Energy Pathways through Reinforcement Learning of Adaptive Steered Molecular Dynamics
Nicholas Ho, John Kevin Cava, John Vant, Ankita Shukla, Jacob Miratsky, Pavan Turaga, Ross Maciejewski, Abhishek Singharoy.
Machine Learning In Structural Biology (MLSB) Workshop at the 36th Conference on Neural Information Processing Systems
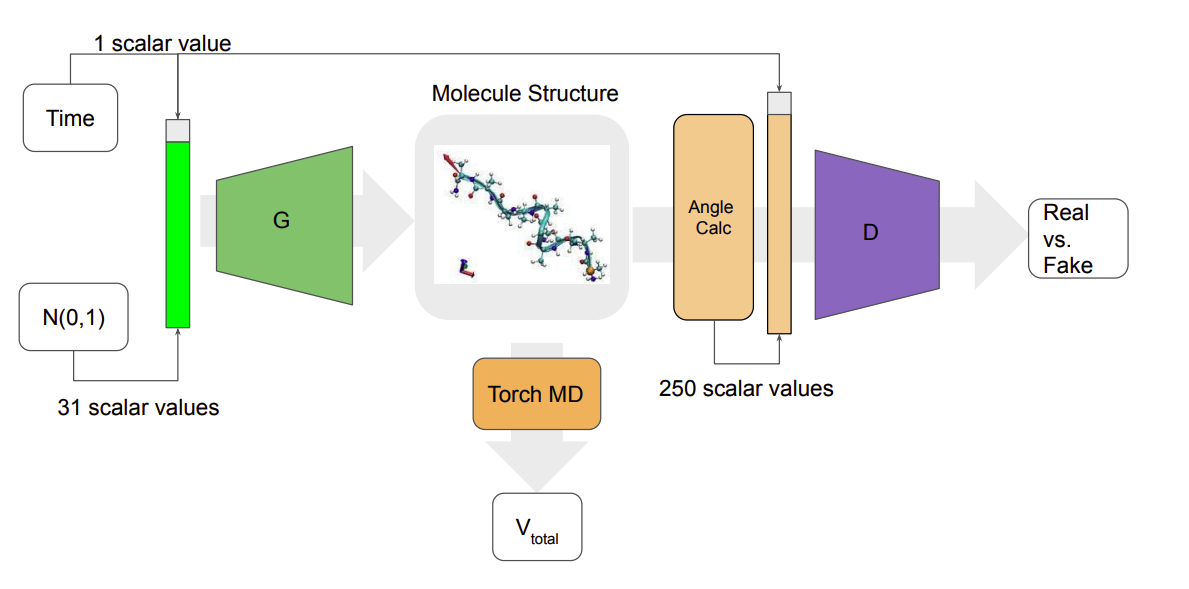
Towards Conditional Generation of Minimal Action Potential Pathways for Molecular Dynamics
John Kevin Cava, John Vant, Nicholas Ho, Ankita Shukla, Pavan Turaga, Ross Maciejewski, Abhishek Singharoy
ELLIS ML4Molecules Workshop, 2021
CyanoPATH: a knowledgebase of genome-scale functional repertoire for toxic cyanobacterial blooms
Wei Du, Gaoyang Li, Nicholas Ho, Landon Jenkins, Drew Hockaday, Jiankang Tan, Huansheng Cao
Briefings in Bioinformatics
Projects
Vitæ
-
Carnegie Mellon University August 2023 - NowPhD Student
Advised by Prof. Jian Ma and Prof. Eric Xing. -
Harvard Medical School June 2022 - May 2023Visiting Research Scholar
• Accepted as a visiting research fellow at Harvard DBMI through a highly competitive application process.
• Currently developing a novel machine learning architecture to deal with missing and imbalanced modalities. -
Struct. Sys. Bio at Biodesign Institute at ASU Feb 2020 - May 2023Research Associate
Worked as a Research Associate in the Singharoy Lab. Lead research for my honors thesis to utilize deep ML methods with statistical mechanical methodologies.
• Lead and wrote a project on discovering free energy pathways with reinforcement learning
• Developed a molecular differentiable simulator for Jax based on TorchMD
• Helped develop novel conditional generative models for deriving free energy pathways
• Implemented and tested several physics-informed models.
-
Carnegie Mellon University June 2021 - August 2021Visiting Research Scholar
Accepted as a visiting research fellow in CMU Statistics through a highly competitive summer research program.
• Created and a novel methodology for using stochastic simulators for soccer game predictions. Our method had comparable results to models trained directly on scores. -
Pichel Lab Biodesign Inst. May 2020 - May 2021Assistant Researcher under Ferran Garcia Pichel at Biodesign Institute
Developed a Python Plugin for Qiime2 for the relationship between ribosomal gene copy number and size. -
Cao Lab Biodesign Inst. August 2019 – May 2021Assistant Researcher
• Conducted data engineering and analysis on genomes assembled from a metagenome in order to study the strain level variance within microbiome communities.
• Implemented a web system that highlighted which genes present in a pathway for particular cyanobacteria species using PFAM’s Hidden Markov Model package, JavaScript, SQL databases, shell and Python. -
Western Tool & Supply June 2020 - July 2020Software Engineer, Intern
• Implemented and trained an LSTM Recurrent Neural Network to predict customer purchase likelihoods. • Integrating and using Bluetooth LE between microcontrollers and Google Chrome into their IOT system. -
TGEN May 2020 - August 2020Virtual Helios Scholar
Due to covid, the Helios Scholars Program was moved to a virtual format. -
Arizona State University August 2019 - May 2023B.Sc. Honors Student
Double Major in Mathematics (4.0) and Computer Science (4.0)
Hobbies
I like to Yoyo!2025 Yoyo Performance (Check this one out)

Thank you Martin Saveski for creating this really neat template!